Hierarchical Sequencing Method - Sequence Tagged Sites
By: HWC
Date Uploaded: 05/29/2020
Tags: homeworkclinic.com Homework Clinic HWC hierarchical sequencing method chromosomes sequence tagged sites bacterial artificial chromosomes genome computer algorithms DNA
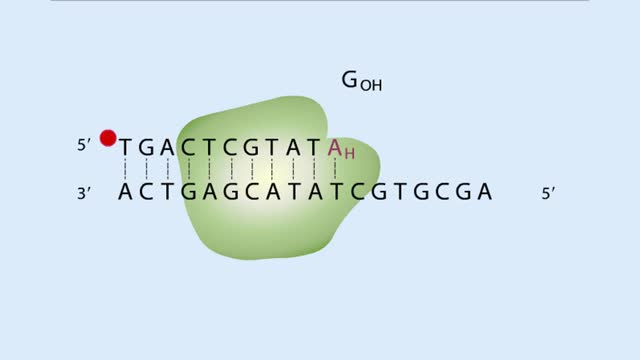
In the hierarchical sequencing method, researchers begin by collecting cells. In humans, each cell contains 23 pairs of chromo-somes. Here we specifically track the DNA from just one of the 23 pairs. Chromosomes have a series of unique DNA sequences, called sequence-tagged sites (STSs), that are already known to researchers. These STSs serve as chromosomal landmarks during later stages of the hierarchical method. The chromosomal DNA is purified, and the other cellular components are removed. The compacted chromosomes uncoil to form long strings of DNA. Enzymes are used to cut the DNA into relatively large fragments, each about 250,000 base pairs (bp) in length. We will focus on the subset of these fragments (indicated in blue) that are derived from the chromosome on the right. The fragments contain the same STSs as the original chromosome. The fragments are cloned into vectors, called bacterial artificial chromosomes, or BACs. The BACs allow the fragments to be replicated in bacteria, which provides enough sample material for the rest of the analysis. Researchers can start from the ends of the vector to identify nearby sequences in the human DNA fragment. If these sequences are unique in the genome, they serve as additional STSs. These STSs can be mapped on the chromosome and used to determine whether the DNA fragments overlap. The hierarchical sequencing method is directed at sequencing these large, overlapping fragments. Yet, before a large fragment can be sequenced, many copies of it are first randomly cut into smaller fragments. After a multistep process of isolating and replicating the small fragments, the DNA of each fragment is sequenced. Although each fragment is sequenced from end to end, we color only small areas to indicate identical sequences among fragments. Computers store the sequence data, and, using computer algorithms, compare the data for overlapping sequences. These common sequences allow the computer to merge information from more than one fragment into one long DNA sequence. Each overlapping large fragment is analyzed until the entire genome sequence is determined.
Add To
You must login to add videos to your playlists.
Advertisement



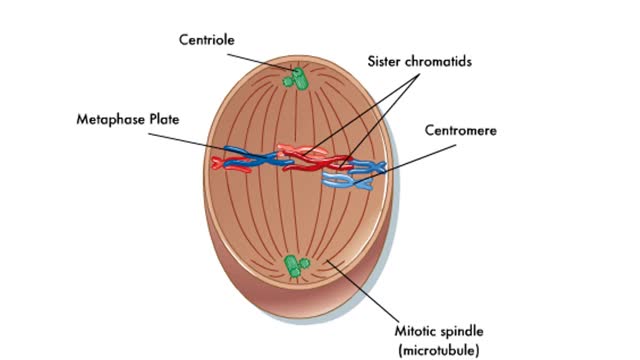
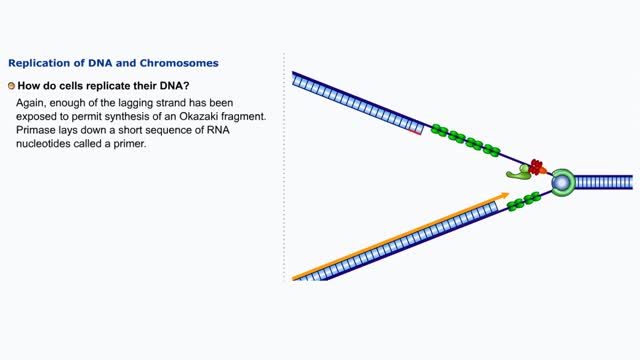
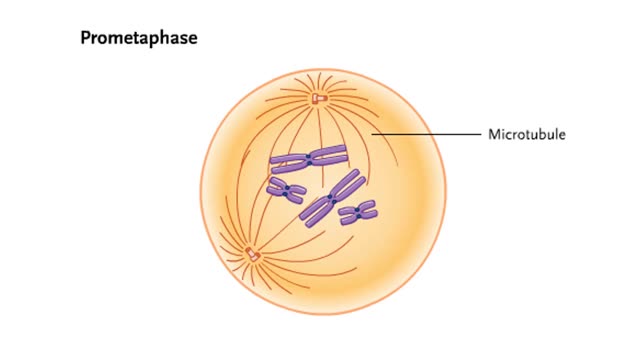
Comments
0 Comments total
Sign In to post comments.
No comments have been posted for this video yet.