ribosome
By: HWC
Date Uploaded: 07/15/2020
Tags: homeworkclinic.com Homework Clinic HWC Trp operon Animation E. coli tryptophan attenuation tryptophan amino acids RNA polymerase DNA ribosome
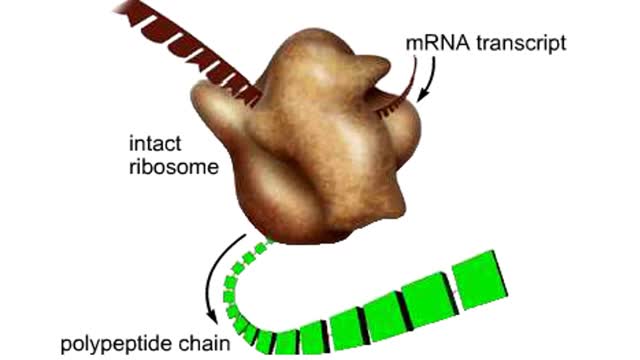
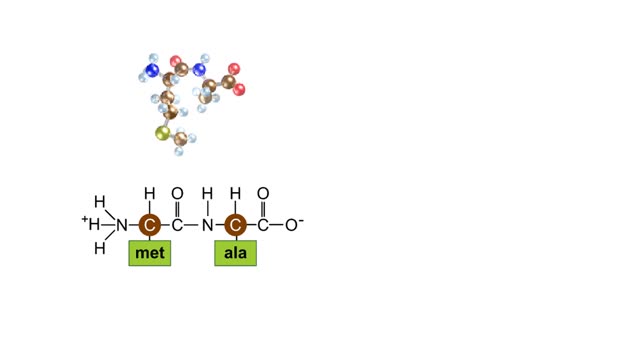
The trp operon in E. coli contains five structural genes corresponding to enzymes that convert chorismate into tryptophan. The trpE gene contains a 5' untranslated region that plays an important role in the regulatory mechanism called attenuation. The 5' UTR contains four regions. Region 1 encodes a short peptide containing tryptophan amino acids. When tryptophan levels are high, region 3 pairs with region 4. This structure terminates, or attenuates, transcription. When tryptophan levels are low, region 2 pairs with region 3. This structure does not terminate transcription. Let's examine how these alternate structures arise. RNA polymerase begins transcribing the DNA. Closely following RNA polymerase, a ribosome begins to translate region 1. When tryptophan is abundant, the ribosome doesn't slow down at the tryptophan codons. By the time region 3 is transcribed, the ribosome has already moved past the tryptophan codons to partly cover region 2. Because region 3 is prevented from pairing with region 2, it pairs with region 4 instead to produce the attenuator, which terminates transcription. The structural genes are not transcribed, and therefore no additional tryptophan is synthesized. When tryptophan is scarce, the ribosome stalls at the tryptophan codons. Therefore, region 2 is not covered when region 3 is transcribed, allowing regions 2 and 3 to form a hairpin that does not terminate transcription. Because region 3 is paired with region 2, it cannot pair with region 4 and form an attenuator. Transcription continues. RNA polymerase transcribes the structural genes, which are translated into enzymes, which in turn synthesize more tryptophan. You can mimic the effect of tryptophan levels by determining how long the ribosome pauses at the Trp codons.
Add To
You must login to add videos to your playlists.
Advertisement



Comments
0 Comments total
Sign In to post comments.
No comments have been posted for this video yet.